Efficient Knowledge Distillation of SAM for Medical Image Segmentation
Medical image segmentation plays a vital role in numerous healthcare applications, such as diagnosing diseases, planning treatments, and guiding surgeries. However, the current gold-standard models for segmentation, like the Segment Anything Model (SAM), often suffer from computational inefficiencies that make them unsuitable for real-time or resource-constrained environments. In this post, we delve into a knowledge distillation approach that significantly improves the computational efficiency of SAM, making it ideal for medical image segmentation tasks in such settings.
Introduction to SAM and Its Challenges
SAM has emerged as a cutting-edge tool for interactive image segmentation. By using a Vision Transformer (ViT) encoder paired with a prompt-guided decoder, SAM achieves remarkable segmentation accuracy across various datasets. However, this performance comes at a high computational cost, making it challenging to deploy in environments with limited resources, like mobile devices or edge computing platforms.
While models like MobileSAM reduce this complexity by using a smaller encoder (ViT-Tiny), they often suffer from degraded segmentation accuracy, especially when tasked with complex applications like medical imaging. This is where the proposed KD SAM approach steps in—by using knowledge distillation to maintain segmentation performance while reducing computational overhead.
Knowledge Distillation: A Solution to Model Complexity
Knowledge distillation is a technique that involves transferring knowledge from a larger, more complex model (the teacher) to a smaller, more efficient one (the student). The goal is to make the student model approximate the performance of the teacher while having fewer parameters and requiring less computation.
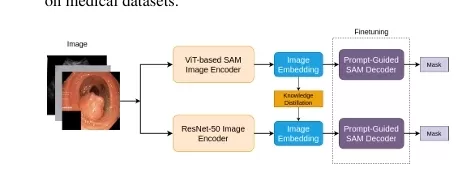
In the case of SAM, we use encoder-decoder knowledge distillation—an approach that optimizes both the encoder and decoder components of the model to ensure high-quality segmentation while significantly reducing the computational cost. By leveraging a dual-loss framework, we combine Mean Squared Error (MSE) loss to capture structural features and Perceptual Loss to preserve semantic details.
The Two-Phase Distillation Process
The KD SAM approach involves a decoupled distillation process, split into two key phases: encoder distillation and decoder fine-tuning.
1. Encoder Knowledge Distillation
The first phase focuses on distilling knowledge from the large ViT encoder in SAM to a more efficient ResNet-50 encoder. The ResNet-50 model is chosen for its balance between model size and performance, making it ideal for resource-constrained environments. The distillation is driven by a combined loss function that includes:
- MSE Loss: This ensures that the student model mimics the low-level structural features of the teacher model.
- Perceptual Loss: This focuses on maintaining high-level semantic details, crucial for accurate segmentation, especially in complex medical imaging tasks.
The MSE loss is calculated on feature maps from the ViT encoder (teacher) and ResNet encoder (student), ensuring structural information is preserved. The Perceptual Loss, derived from pre-trained VGG networks, further ensures that the student model captures semantic features, which are key for distinguishing complex structures in medical images.
2. Decoder Fine-Tuning
In the second phase, we fine-tune the decoder to work effectively with the distilled encoder. Unlike traditional methods, where the encoder and decoder are trained simultaneously, our decoupled approach trains the encoder first and adapts the decoder to work with the new encoder feature representations. For fine-tuning, we use Dice Loss, which is particularly well-suited for medical segmentation tasks due to its ability to handle class imbalance—a common issue in medical datasets.
Experimental Setup
The effectiveness of KD SAM is demonstrated on several medical image segmentation datasets:
- Kvasir-SEG: A gastrointestinal polyp segmentation dataset.
- ISIC 2017: A melanoma skin cancer dataset.
- Fetal Head Ultrasound: Used to segment fetal brain structures in ultrasound images.
- Breast Ultrasound: Used for identifying and segmenting regions in breast ultrasound images.
The model is trained in two phases:
- Encoder distillation: The ResNet-50 model is trained using a combined MSE and Perceptual Loss function to distill knowledge from the ViT encoder.
- Decoder fine-tuning: The SAM decoder is fine-tuned with Dice Loss, focusing on optimizing segmentation accuracy without overburdening the system.
Results and Performance
The results demonstrate that KD SAM outperforms traditional models like MobileSAM, achieving comparable or even superior segmentation accuracy while maintaining significantly lower computational costs. Some key findings include:
- Comparable segmentation accuracy on challenging datasets like ISIC 2017 and Kvasir-SEG.
- Reduced model size and faster inference time, making it more suitable for deployment in real-time applications.
- Improved performance on complex medical images, where fine-grained segmentation is essential.
Conclusion
The KD SAM model provides an efficient solution for real-time medical image segmentation by leveraging knowledge distillation to retain high accuracy while dramatically reducing computational overhead. By targeting both the encoder and decoder optimization, KD SAM balances segmentation performance and computational efficiency, making it well-suited for deployment in resource-constrained environments such as mobile devices and edge computing platforms.
This work presents an innovative approach that not only improves medical image segmentation but also holds promise for broader applications in interactive image segmentation tasks across industries.
Discussion Prompt: How do you think distillation techniques, like those used in KD SAM, will influence the future of real-time medical image processing on edge devices?
What's Your Reaction?